Evolution

Evolution is the change of replicating entities through time due to descent with modification. This topic includes projects modelling the fundamental processes and forces that drive evolution itself, as well as projects that make use of evolutionary theory to make predictions and inferences about current organisms and biomolecules, particularly DNA and protein. This topic also covers Population Genetics. Evolution is not to be confused with morphogenesis, which can be a gradual change with time but does not involve replication.
For queries about this topic, contact Richard Edwards.
View the calendar of events relating to this topic.
Projects

A novel approach to analysing fixed points in complex systems
James Dyke (Investigator), Iain Weaver
This work aims to contribute to our understanding of the relationship between complexity and stability. By describing an abstract coupled life-environment model, we are able to employ novel analytical, and computational techniques to shed light on the properties of such a system.

Associative learning in ecosystems: Network level adaptation as an emergent propery of local selection
Richard Watson, James Dyke (Investigators), Daniel Power
Ecosystems may exhibit collective adaptive properties that arise from natural selection operating on their component species. These properties include the ability of the ecosystem to return to specific configurations of species, in a manner highly analogous to mechanisms of associative learning in neural networks.
Benchmarking the GOPHER orthologue prediction algorithm.
Richard Edwards, Shaun Maguire
Generation of Orthologous Proteins from High-throughput Evolutionary Relationships (GOPHER) is an orthologue prediction algorithm. This experiment aims to benchmark this algorithm.
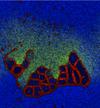
Cellular Automata Modelling of Membrane Formation and Protocell Evolution
Seth Bullock (Investigator), Stuart Bartlett
We simulated the meso-level behaviour of lipid-like particles in a range of chemical and physical environments. Self-organised protocellular structures can be shown to emerge spontaneously in systems with random, homogeneous initial conditions. Introducing an additional 'toxic' particle species and an associated set of synthesis reactions produced a new set of ecological behaviours compared to the original model of Ono and Ikegami.

Centre for Doctoral Training in Next Generation Computational Modelling
Hans Fangohr, Ian Hawke, Peter Horak (Investigators), Susanne Ufermann Fangohr, Thorsten Wittemeier, Kieran Selvon, Alvaro Perez-Diaz, David Lusher, Ashley Setter, Emanuele Zappia, Hossam Ragheb, Ryan Pepper, Stephen Gow, Jan Kamenik, Paul Chambers, Robert Entwistle, Rory Brown, Joshua Greenhalgh, James Harrison, Jonathon Waters, Ioannis Begleris, Craig Rafter
The £10million Centre for Doctoral Training was launched in November 2013 and is jointly funded by EPSRC, the University of Southampton, and its partners.
The NGCM brings together world-class simulation modelling research activities from across the University of Southampton and hosts a 4-year doctoral training programme that is the first of its kind in the UK.

Development and application of powerful methods for identifying selective sweeps
Andrew Collins, Reuben Pengelly, Timothy Sluckin, Sarah Ennis (Investigators), Clare Horscroft
This project is about detecting regions of the genome which have experienced selective pressure. To achieve this, mathematical models will be developed and applied to human genomic data sets, as well as to those of other species.

Differences between sexual genetic algorithms and a compositional cooperative co-evolutionary algorithm
Richard Watson (Investigator), Daniel Power
Different algorithms are representative of different genetic processes. This work explores how algorithms representing sexual recombination can solve certain problems that hill climbers cannot.

Do the adaptive dynamics of host-parasite systems catalyse or constrain sympatric speciation?
Richard Watson (Investigator), Daniel Power
Coevolutionary dynamics affect both parties evolutionary trajectories. When might these affect speciation? This project uses a simulation model to explore the issue.
Identification of novel Crustacean Pathogen Receptor Proteins
Richard Edwards, Chris Hauton, Timothy Elliott (Investigators), Oyindamola Lawal, Lloyd Mushambadzi
We are mining EST libraries (sequence fragments of expressed genes) for novel proteins that might play a role in the immune response of crustaceans.
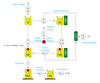
Immunotherapy Research: Modelling MHC Class I Complex Assembly
Timothy Elliott, Jorn Werner (Investigators), Alistair Bailey
This project uses mathematical modelling and simulation to investigate mechanisms by which our cells process and present biological information that is used by our immune system to distinguish between healthy and diseased cells.

Impact of reciprocal feedbacks between evolution and ecology
Richard Watson, Patrick Doncaster, James Dyke (Investigators), Daniel Power
How do organism's activities affect their evolutionary trajectories? This project uses simulation techniques to evaluate the effects of this feedback.

Integrated in silico prediction of protein-protein interaction motifs
Richard Edwards (Investigator), Nicolas Palopoli, Kieren Lythgow
Many vital protein-protein interactions are mediated by Short Linear Motifs (SLiMs) which are short proteins typically 5-15 amino acids long containing only a few positions crucial to function. This project integrates a number of leading computational techniques to predict novel SLiMs and add crucial detail to protein-protein interaction networks.

Interactome-wide prediction of short linear protein interaction motifs in humans
Richard Edwards (Investigator)
Short Linear Motifs (SLiMs) are important in many protein-protein interactions. In previous work, we have developed a computational tool, SLiMFinder, which places the interpretation of evidence for motifs within a statistical framework with high specificity, and subsequently enhanced sensitivity through application of conservation-based sequence masking. We are now applying these tools to a comprehensive set of human protein-protein interactions in order to predict novel human SLiMs in silico.

Mass Spec identification of proteins utilising EST libraries
Richard Edwards, Maria Debora Iglesias-Rodriguez (Investigators), Bethan Jones
Expressed Sequence Tag (EST) data presents a particular challenge for the identification of proteins using mass spectrometry (MS): it is often redundant (multiple copies of the same gene), consists primarily of short fragments of coding sequence, contains many sequencing errors and is generally poorly annotated. We are developing computational pipelines to maximise robust protein identifications from EST data despite these challenges.

Origins of Evolvability
Richard Watson, Markus Brede (Investigators), William Hurndall
This project examined the putative evolvability of a Lipid World model of fissioning micelles. It was demonstrated that the model lacked evlovability due to poor heritability. Explicit structure for micelles was introduced along with a spatially localised form of catalysis which increased the strength of selection as coupling between potential chemical units of heredity were reduced.

Perceived Attractiveness as a Factor Affecting Condomless Sex
Anastasia Eleftheriou, Seth Bullock
The objectives of this project are to better understand the relationship between perceived attractiveness and condom use intentions and to gain insight into the relationship between perceived attractiveness and potential sexual risk behaviours.

Selection pressure for language and theory-of-mind in monkeys
Jason Noble (Investigator)
To what extent are the alarm calls of putty-nosed monkeys likely to be a good model for human language evolution? Simulation is used to classify evolutionary trajectories as either plausible or implausible, and to put lower bounds on the cognitive complexity required to perform particular behaviours.

The Origins of Communication Revisited
Jason Noble (Investigator), Jordi Arranz
Quinn (2001) sought to demonstrate that communication be- tween simulated agents could be evolved without pre-defined communication channels. Quinn’s work was exciting because it showed the potential for ALife models to look at the real origin of communication; however, the work has never been replicated. In order to test the generality of Quinn’s result we use a similar task but a completely different agent architecture. We find that qualitatively similar behaviours emerge, but it is not clear whether they are genuinely communicative. We extend Quinn’s work by adding perceptual noise and internal state to the agents in order to promote ritualization of the nascent signal. Results were inconclusive; philosophical implications are discussed.

The Social-cognitive Niche: An Exploration of the Co-evolutionary Relationship between Human Mind and language, with a Particular Focus of the Self-organisational properties of the Emergence of Symbolic Representation.
Jason Noble, Glyn Hicks (Investigators), Lewys Brace
This work explored the relationship between the origin and subsequent evolution of the human mind and language; a relationship that is believed to be symbiotic in nature. This piece aimed to achieve two objectives. Firstly, it set out a theoretical framework, using the principles of complexity theory and self-organisation, which attempts to explain this relationship from a holistic perspective.
Secondly, it presented an agent-based model of a vervet monkey social group, which sought to investigate the variables that were perceived to underpin the emergence of symbolic representation within a population of language users.
The belief here was that, by understanding the influence of these variables, one would be able to better understand the genesis of the aforementioned relationship.

µ-VIS Computed Tomography Centre
Ian Sinclair, Richard Boardman, Dmitry Grinev, Philipp Thurner, Simon Cox, Jeremy Frey, Mark Spearing, Kenji Takeda (Investigators)
A dedicated centre for computed tomography (CT) at Southampton, providing complete support for 3D imaging science, serving Engineering, Biomedical, Environmental and Archaeological Sciences. The centre encompasses five complementary scanning systems supporting resolutions down to 200nm and imaging volumes in excess of one metre: from a matchstick to a tree trunk, from an ant's wing to a gas turbine blade.
People
 Seth Bullock
Seth BullockProfessor, Electronics and Computer Science (FPAS)
 Andrew Collins
Andrew CollinsProfessor, Medicine (FM)
 Simon Cox
Simon CoxProfessor, Engineering Sciences (FEE)
 Timothy Elliott
Timothy ElliottProfessor, Medicine (FM)
 Sarah Ennis
Sarah EnnisProfessor, Medicine (FM)
 Hans Fangohr
Hans FangohrProfessor, Engineering Sciences (FEE)
 Jeremy Frey
Jeremy FreyProfessor, Chemistry (FNES)
 Ian Sinclair
Ian SinclairProfessor, Engineering Sciences (FEE)
 Timothy Sluckin
Timothy SluckinProfessor, Mathematics (FSHS)
 Mark Spearing
Mark SpearingProfessor, Engineering Sciences (FEE)
 Patrick Doncaster
Patrick DoncasterReader, Biological Sciences (FNES)
 Peter Horak
Peter HorakReader, Optoelectronics Research Centre
 Tiina Roose
Tiina RooseReader, Engineering Sciences (FEE)
 Jorn Werner
Jorn WernerReader, Biological Sciences (FNES)
 Markus Brede
Markus BredeSenior Lecturer, Electronics and Computer Science (FPAS)
 Stuart Clarke
Stuart ClarkeSenior Lecturer, Medicine (FM)
 Reuben Pengelly
Reuben PengellySenior Lecturer, Medicine (FM)
 Fraser Sturt
Fraser SturtSenior Lecturer, Humanities (FH)
 Richard Watson
Richard WatsonSenior Lecturer, Electronics and Computer Science (FPAS)
 Christopher Bell
Christopher BellLecturer, Biological Sciences (FNES)
 Srinandan Dasmahapatra
Srinandan DasmahapatraLecturer, Electronics and Computer Science (FPAS)
 James Dyke
James DykeLecturer, Electronics and Computer Science (FPAS)
 Ian Hawke
Ian HawkeLecturer, Mathematics (FSHS)
 Glyn Hicks
Glyn HicksLecturer, Humanities (FH)
 Maria Debora Iglesias-Rodriguez
Maria Debora Iglesias-RodriguezLecturer, Ocean & Earth Science (FNES)
 Philipp Thurner
Philipp ThurnerLecturer, Engineering Sciences (FEE)
 Richard Boardman
Richard BoardmanSenior Research Fellow, Engineering Sciences (FEE)
 Chris Hauton
Chris HautonSenior Research Fellow, Ocean & Earth Science (FNES)
 Alistair Bailey
Alistair BaileyResearch Fellow, Medicine (FM)
 Dmitry Grinev
Dmitry GrinevResearch Fellow, Engineering Sciences (FEE)
 Rob Mills
Rob MillsResearch Fellow, Electronics and Computer Science (FPAS)
 Jason Noble
Jason NobleResearch Fellow, Electronics and Computer Science (FPAS)
 - -
- -Postgraduate Research Student, Electronics and Computer Science (FPAS)
 Jordi Arranz
Jordi ArranzPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Alice Ball
Alice BallPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Stuart Bartlett
Stuart BartlettPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Ioannis Begleris
Ioannis BeglerisPostgraduate Research Student, Engineering Sciences (FEE)
 Harry Beviss
Harry BevissPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Lewys Brace
Lewys BracePostgraduate Research Student, Electronics and Computer Science (FPAS)
 Rory Brown
Rory BrownPostgraduate Research Student, Civil Engineering & the Environment (FEE)
 Jamie Caldwell
Jamie CaldwellPostgraduate Research Student, Engineering Sciences (FEE)
 Paul Chambers
Paul ChambersPostgraduate Research Student, Engineering Sciences (FEE)
 Caroline Duignan
Caroline DuignanPostgraduate Research Student, Biological Sciences (FNES)
 Anastasia Eleftheriou
Anastasia EleftheriouPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Graham Elliott
Graham ElliottPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Robert Entwistle
Robert EntwistlePostgraduate Research Student, Engineering Sciences (FEE)
 Greg Fisher
Greg FisherPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Stephen Gow
Stephen GowPostgraduate Research Student, Engineering Sciences (FEE)
 Joshua Greenhalgh
Joshua GreenhalghPostgraduate Research Student, Engineering Sciences (FEE)
 James Harrison
James HarrisonPostgraduate Research Student, Engineering Sciences (FEE)
 Tom Hebbron
Tom HebbronPostgraduate Research Student, Electronics and Computer Science (FPAS)
 William Hurndall
William HurndallPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Adam Jackson
Adam JacksonPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Guy Jacobs
Guy JacobsPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Bethan Jones
Bethan JonesPostgraduate Research Student, National Oceanography Centre (FNES)
 Konstantinos Kouvaris
Konstantinos KouvarisPostgraduate Research Student, Electronics and Computer Science (FPAS)
 David Lusher
David LusherPostgraduate Research Student, Engineering Sciences (FEE)
 Alvaro Perez-Diaz
Alvaro Perez-DiazPostgraduate Research Student, Engineering Sciences (FEE)
 Can Pervane
Can PervanePostgraduate Research Student, Electronics and Computer Science (FPAS)
 Daniel Power
Daniel PowerPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Craig Rafter
Craig RafterPostgraduate Research Student, Engineering Sciences (FEE)
 Hossam Ragheb
Hossam RaghebPostgraduate Research Student, Engineering Sciences (FEE)
 Sonya Ridden
Sonya RiddenPostgraduate Research Student, Mathematics (FSHS)
 Sabin Roman
Sabin RomanPostgraduate Research Student, University of Southampton
 Iza Romanowska
Iza RomanowskaPostgraduate Research Student, Humanities (FH)
 Kieran Selvon
Kieran SelvonPostgraduate Research Student, Engineering Sciences (FEE)
 Ashley Setter
Ashley SetterPostgraduate Research Student, Engineering Sciences (FEE)
 Nathan Smith
Nathan SmithPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Nick Synes
Nick SynesPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Simon Tudge
Simon TudgePostgraduate Research Student, Electronics and Computer Science (FPAS)
 Jonathon Waters
Jonathon WatersPostgraduate Research Student, Engineering Sciences (FEE)
 Iain Weaver
Iain WeaverPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Thorsten Wittemeier
Thorsten WittemeierPostgraduate Research Student, Engineering Sciences (FEE)
 Emanuele Zappia
Emanuele ZappiaPostgraduate Research Student, Engineering Sciences (FEE)
 Davide Zilli
Davide ZilliPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Shaun Maguire
Shaun MaguireUndergraduate Research Student, Biological Sciences (FNES)
 Elena Vataga
Elena VatagaTechnical Staff, iSolutions
 Petrina Butler
Petrina ButlerAdministrative Staff, Research and Innovation Services
 Susanne Ufermann Fangohr
Susanne Ufermann FangohrAdministrative Staff, Civil Engineering & the Environment (FEE)
 Richard Edwards
Richard EdwardsAlumnus, University of New South Wales, Australia
 Jan Kamenik
Jan KamenikAlumnus, University of Southampton
 Oyindamola Lawal
Oyindamola LawalAlumnus, former UG, Biological Sciences
 Kieren Lythgow
Kieren LythgowAlumnus, Health Protection Agency
 Lloyd Mushambadzi
Lloyd MushambadziAlumnus, former UG, Biological Sciences
 Nicolas Palopoli
Nicolas PalopoliAlumnus, Biological Sciences (FNES)
 Kenji Takeda
Kenji TakedaAlumnus, Engineering Sciences (FEE)
 Brian Bonney
Brian BonneyNone, None
 Clare Horscroft
Clare HorscroftNone, None