Lagrangian modelling of ecosystem dynamics at the Bermuda Atlantic Time-series Study station
- Started
- 1st October 2010
- Research Team
- Melissa Saeland
- Investigators
- Tom Anderson, Seth Bullock
Research area
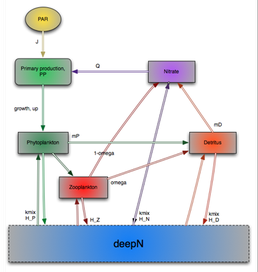
Concerns surrounding the changing climate have created a need to understand marine ecosystem dynamics, as this is central to the study of the ocean’s role in the global carbon cycle. The oligotrophic gyres of the ocean are of great scientific interest, as they are so vast, and the Bermuda Atlantic Time series site (BATS) has been intensively studied over 20 years, and so is ideal for studying these areas. Previous modelling studies (e.g. Anderson and Pondaven 2003; Salihoglu et al, 2008) have used Eulerian models (in which different plankton groups are aggregated into state variables that are assumed to be homogeneous within each model layer) to study nutrient cycling, community structure and export at the BATS site. Focus is starting to move to Lagrangian models, which involve the plankton being represented as individuals, allowing for individual variability to be represented.
Research question
There exists uncertainty around how the increase in atmospheric carbon dioxide will affect the plankton ecosystems in the ocean and biogeochemical ecosystem models can be a valuable tool when trying to make predictions surrounding this area of science. The aim of the project is to investigate the suitability of Lagrangian models for representing ecosystems as well as to try and create a stable model that successfully reproduces the marine plankton ecosystem at the BATS site.
Research methodology
The first part of the PhD will build on from the summer project, which investigated the implementation of Lagrangian models. It will utilise different software packages, such as NetLogo and the Virtual Ecology Workbench (VEW) to investigate the applicability and potential advantages / disadvantages of Lagrangian models for the simulation of plankton ecosystems. In the second part of the project, the Virtual Ecology Workbench (VEW) will be used for setting up, parametersing and running a model at the BATS site. VEW is a complex piece of plankton ecosystem modelling software, specifically designed for agent-based, Lagrangian simulations. There is currently little research that has been carried out with this particular piece of software, so the initial model will be very simple and will be developed in tandem with an identical model in NetLogo, to check the validity of the software. Once an appropriate model is constructed, the initial simple representation can be expanded to separate functional groups, such as picoplankton, nitrogen fixers and diatoms, distinguish micro- and mesozooplankton and also include bacteria and dissolved organic matter. The output of the model can then be compared to field data taken from the BATS station. The role of different organisms and processes and how they contribute to processes in biogeochemical cycling and export can then be assessed.
Categories
Life sciences simulation: Ecology
Algorithms and computational methods: Agents
Simulation software: NetLogo
Programming languages and libraries: Fortran, Python
Computational platforms: Mac OS X
Transdisciplinary tags: Complex Systems, Quantitative Biology, Scientific Computing