Investigation into the Interfacial Physics of Field Effect Biosensors
- Started
- 7th October 2013
- Research Team
- Benjamin Lowe
- Investigators
- Nicolas Green, Chris-Kriton Skylaris
This interdisciplinary research aims to improve understanding of Field Effect Transistor Biosensors (Bio-FETs) and to work towards a multiscale model which can be used to better understand and predict device response.
Recent advances in nanotechnology have led to the development of a large range of electronic devices with applications across the physical, chemical and biological disciplines. Biosensors represent a class of electronic devices which involve a complex interplay between aspects of all these disciplines. Field Effect Transistor biosensors are an active area of research within Southampton as they have been shown to be capable of detecting single molecule binding. Their low-cost and simple fabrication process means that these sensors mean that they have great potential as, for example, point-of-care healthcare devices; however their potential as electrochemical transistors may extend well beyond biosensors.
The impressive sensitivity of the devices arises because of direct interaction between the internal electrical behaviour of the device and the short range electrostatic/electrochemical forces generated by molecules both in suspension and during binding to sensing sites on the device. The driving force for this phenomena, known as the field-effect, is well known and is the mechanism of transistors present in modern microprocessors. In contrast to field-effect transistors, this system is electrochemical and involves molecular scale interactions which significantly increase the complexity of the system, introducing new dynamics on various length- and time- scales, and, as a result, the interfacial physics remains poorly understood. To span the relevant time- and length-scales, the modelling of these systems will require an integration of disparate methods.
The project aims towards three distinct but related objectives:
-
Improved understanding of the mechanism of detection in these systems and therefore some investigation into ion dynamics and the electric double layer.
-
To improve rational design of devices by incorporating novel molecular-scale insight into the relevant interfacial physics. For example, suggesting changes to device dimensions for improved sensitivity. Advances in this field have direct industrial and scientific relevance.
The project will aim to work synergistically with the existing researchers working on the fabrication and testing of Silicon Nanowire Biosensors within Southampton's Nanofabrication Centre.
The precise mechanism of Field Effect Sensors is still poorly understood, however it is known that the response of the system is affected by a range of factors, such as nanowire size, surface composition and pH. Due to the complexity of the system, many of the previous attempts to predict the response of devices have focused a qualitative description of the system, predicting only the direction of the signal change based on the charge on the biomolecule and the doping of the nanowire. Previous attempts to quantitatively describe the response of the biosensor have all been based on solving the mean-field Poisson-Boltzmann equation, but have neglected an explicit description of the ion dynamics and polarisation effects which may be vital to understanding the mechanism of operation.
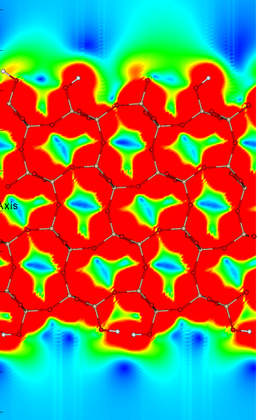
Below is an example of an Ab Initio Molecular Dynamics (AIMD) simulation calculated as part of this work[1]:
I am collaborating with Yuki Maekawa, Yasushi Shibuta and Toshiya Sakata at the University of Tokyo to understand interfacial ion dynamics at the surface of BioFETs. Paper submitted for publication.
Publications:
[1] Lowe, B. M., Skylaris, C. -K., Green. N. G. Acid-Base Dissociation Mechanisms and Energetics at the Silica-Water Interface: An Activationless Process. Journal of Colloid and Interface Science. 451 (2015) 231–244. http://dx.doi.org/10.1016/j.jcis.2015.01.094
Categories
Life sciences simulation: Biomolecular simulations, Nanoscale Assemblies
Physical Systems and Engineering simulation: Biometrics, Complex fluids, Diffusion, Electromagnetism, Materials, Semiconductors, Sensors
Algorithms and computational methods: Density functional Theory, Finite elements, Kinetic Monte Carlo, Molecular Dynamics, Molecular Mechanics, Monte Carlo, Multi-physics, Multi-scale, Quantum Chemistry, Quantum Computation, statistical analysis
Simulation software: CASTEP, COMSOL, NWCHEM, Onetep
Visualisation and data handling software: Mayavi, VisIt, VMD
Software Engineering Tools: Vim
Programming languages and libraries: Python
Computational platforms: Iridis, Linux, Windows
Transdisciplinary tags: Complex Systems, HPC, Scientific Computing