Whole exome sequencing identifies novel FLNA mutation in familial Ebstein's anomaly
- Research Team
- Gaia Andreoletti
- Investigators
- Jane Gibson, Andrew Collins, Sarah Ennis
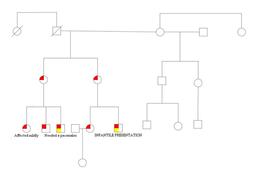
Family pedigree of family of which 8 individuals are affected by Ebstein's anomaly.Pedigree symbols: red filled symbol indicates affected individual;yellow filled symbol indicates patients exome sequenced.
Ebstein's anomaly is a rare congenital heart disease which accounts for less than 1% of all congenital heart diseases . In 1991 Keeton and collaborators reported a family in which eight people in three generations had Ebstein's anomaly. To understand the genetics behind this familial hereditary disease, two first cousins of the third generation underwent whole exome sequencing .
This study aims to identify the shared rare, evolutionarily deleterious, and functional variations between the two cousins by interrogating the exome data for a set of 193 genes previously involved in heart defects.
Genomic DNA was extracted using the salting out method and the samples were sent to be sequenced at the Wellcome Trust Centre for Human Genetics in Oxford. Through Iridis, the large amount of raw data generated, about 30 gigabases of DNA sequence per exome, were aligned to the human reference genome (hg19) with Novoalign and variant calling was performed with Samtools. Quality control tests were made using customised scripts. The gene list was obtained by using the January 2014 version of the Human Gene Mutation Database.
Across the panel of genes the two cousins share a novel non-synonymous G>A highly conserved and deleterious variants in filamin A.
FLNA is a gene previously involved in cardiac conditions but not in Ebstein’s. The mutations found cause a truncation of the protein structure altering the protein function. Although functional studies are required to fully understand the molecular mechanisms of the mutation, this study suggests FLNA as a novel candidate gene for Ebstein’s anomaly.
Categories
Life sciences simulation: Bioinformatics, Epidemiology, NextGen Sequencing
Programming languages and libraries: Perl