The autotransporter ? domain: insights into structure and function through multi scale molecular dynamics simulations
- Started
- 10th January 2011
- Ended
- 1st October 2012
- Research Team
- Daniel Holdbrook, Thomas Piggot
- Investigators
- Syma Khalid
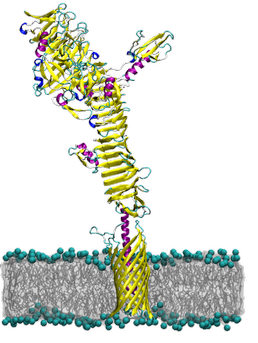
The autotransporters are a family of bacterial outer membrane proteins that are characterised by three functional domains: an N-terminal signal sequence, a central passenger domain, and a C-terminal transmembrane ?-barrel domain. Using multi-scale molecular dynamics simulations we explore the structural dynamics and preferred membrane localisation of the ?-barrel domains from five autotransporters: NalP, EspP, EstA, Hia and Hbp. The ?-barrel domain has been proposed to aid in the translocation of the passenger domain across the outer membrane. In addition, some autotransporters have been shown to localise to the poles of rod shaped bacteria.
We employ atomistic molecular dynamics simulations to investigate the flexibility of the ?-barrel domains from the five autotransporters in lipids bilayers designed to capture much of the complexity of the bacterial outer membrane. Furthermore, we explore the structure-function relationship of EstA from Pseudomonas aeruginosa by simulating the translocator domain and the associated passenger domain. We employ coarse-grained molecular dynamics simulations to investigate the membrane localisation and potential preference of these proteins for specific lipid types. Thus the combined atomistic and coarse grained approach allows us to explore long time and length scales, adding detail, where necessary.
Categories
Life sciences simulation: Structural biology