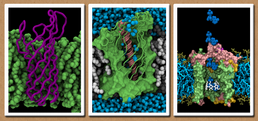
Dr Syma Khalid
- Position
- Principal Research Fellow
- Institution
- Chemistry (FNES)
- Webpage
- http://www.personal.soton.ac.uk/sk2x07
- Telephone
- 02380 594176
- S.Khalid@soton.ac.uk
- Contact
- Complete this online contact form to contact Syma.
Research
The systems and synthetic biology modelling group, headed by Dr Syma Khalid is based in the Chemical Biology sub-section of the School of Chemistry, University of Southampton. Our research involves the application of computational techniques to the study of large biological molecules and systems including bacterial outer membrane proteins and nucleic acids. One of our key areas of research involves applying the knowledge gained from studying biological systems to aid the design of synthetic systems for applications in bionanotechnology. We are particularly interested in developing methods to bridge the gap between traditional biomolecular simulation and systems biology.
We have strong links with experimental groups in physical biology, biochemistry, chemical biology and structural biology
Current Projects
Bacterial Outer Membrane Proteins
Stochastic Sensors
Stochastic biosensors are built around a membrane containing a cylindrically shaped protein (usually a bacterial outer membrane protein). A voltage applied across the membrane generates an ionic current that pulls charged analyte molecules through the pore. As the analyte molecule is pulled through the nanopore, it blocks the current by a characteristic amount, thus leaving a trace of itself. The characteristic decrease in the current arising from differences in their chemical properties allows discrimination between different analyte molecules. We are undertaking simulation studies of bacterial proteins and model nanopores based on proteins, to aid the development of robust and efficient biosensors suitable for the detection of a variety of analytes. This project is funded through a BBSRC CASE award with Oxford Nanopore Technologies as the industrial partner.
Linking Structure to Function
We are interested in the link between the structure of bacterial outer membrane proteins (OMPs) and their function. To this end we have studied the conformational dynamics and interactions with membranes of a variety of OMPs.
DNA-lipid interactions
The E.coli membrane: towards a systems description This project includes the OMSys collaboration with Prof Mark Sansom's group, Oxford
The cohesin hinge region Collaborative project with Professor Kim Nasmyth's group, Oxford
Research Interests
Life sciences simulation: Bioinformatics, Biomolecular Organisation, Biomolecular simulations, Microbiology, Nanoscale Assemblies, Structural biology, Systems biology
Physical Systems and Engineering simulation: Biomechanics, Elasticity, Structural dynamics
Algorithms and computational methods: FFT, Molecular Dynamics, Molecular Mechanics
Visualisation and data handling methods: Database
Simulation software: Gromacs
Visualisation and data handling software: Gnuplot, MS Office Access, VMD, Xmgrace
Software Engineering Tools: CVS, Emacs
Programming languages and libraries: C, Fortran, Perl, Python, Tcl
Computational platforms: HECToR, HPCx, Iridis, Linux, Mac OS X
Transdisciplinary tags: Complex Systems, Computer Science, e-Research, HPC, Scientific Computing, Visualisation
Syma's team members
 James Graham James GrahamPostgraduate Research Student, Electronics and Computer Science (FPAS) |
 Jamie Parkin Jamie ParkinPostgraduate Research Student, Chemistry (FNES) |
 Ric Gillams Ric GillamsPostgraduate Research Student, Chemistry (FNES) |
 Andrew Guy Andrew GuyPostgraduate Research Student, Chemistry (FNES) |
 Josephine Corsi Josephine CorsiAlumnus, University of Southampton |
Joint projects with...
 Philip Williamson Philip WilliamsonSenior Research Fellow, Biological Sciences (FNES) |
Research Groups
Institute for Complex Systems Simulations (ICSS)
University of Southampton
Projects
Lyotropic phase transitions of lipids studied by CG MD simulation and experimental techniques
With Josephine Corsi

Probing the oligomeric state and interaction surface of Fukutin Transmembrane Domain in lipid bilayer via Molecular Dynamics simulations
With Philip Williamson (Investigator), Daniel Holdbrook, Jamie Parkin, Nils Berglund, Yuk Leung
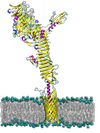
The autotransporter ? domain: insights into structure and function through multi scale molecular dynamics simulations

THE NORM MATE TRANSPORTER FROM N. GONORRHEAE: INSIGHTS INTO DRUG & ION BINDING FROM ATOMISTIC MOLECULAR DYNAMICS SIMULATIONS

Using Molecular Dynamics to Understand the Antibacterial Mechanisms of Daptomycin & Chlorhexidine to Target the Bacterial Membrane
With Eilish McBurnie