Lyotropic phase transitions of lipids studied by CG MD simulation and experimental techniques
- Started
- 24th November 2008
- Research Team
- Josephine Corsi
- Investigators
- Syma Khalid
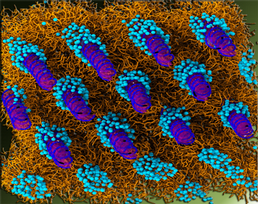
A study of the phase behaviour of cationic lipid - DNA complexes such as those used for transfection by coarse grained molecular dynamics simulation. Lipid systems studied include DOPE, DOPE/DNA and DOPE/DOTAP/DNA. Structural parameters and phase behaviour observed computationally have been compared with those gained using Small Angle X-ray Scattering (SAXS) and polarising light microscopy techniques.
Categories
Life sciences simulation: Biomolecular Organisation, Nanoscale Assemblies
Algorithms and computational methods: Molecular Dynamics
Simulation software: Gromacs