Using Molecular Dynamics to Understand the Antibacterial Mechanisms of Daptomycin & Chlorhexidine to Target the Bacterial Membrane
- Started
- 10th October 2016
- Research Team
- Eilish McBurnie
- Investigators
- Syma Khalid
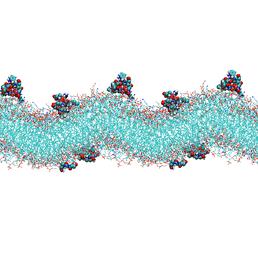
3 Daptomycin Molecules in Staphylococcus aureus Membrane after 1500ns Simulation on Iridis 4 - visualised using VMD
Dependence on antimicrobial agents to treat simple to extremely complex illnesses is ever growing and thus so is bacterial antibiotic resistance. Due to this it is integral to truly understand, on a molecular level how an antibiotics kills a target cell. By understanding the mode of action of an antibiotic, it could not only mean that the drug could be better used in regards to usage and the infections it should be used against, but also give an insight into potential novel antibiotics. Whilst antibiotics kill or prevent growth of bacteria via different several methods; all antibiotics must meet and cross the bacterial membrane meaning understanding this interaction is vital. This is especially true in cases where the antimicrobial kills cells by disrupting the cell membrane, such as with daptomycin and chlorhexidine focused upon in this project. Whilst daptomycin is an antibiotic, which works on specifically gram-positive bacteria, such as Staphylococcus aureus, chlorhexidine not targets both gram positive and negative bacteria.
Based on this growing pressure on antibiotics and the known importance of the cell membrane's interaction with these antimicrobial peptides; this study aims to use computational methods, specifically molecular dynamics using the classic calculations from this method, to understand how daptomycin interferes with the gram positive membrane and how chlorhexidine does so with both gram positive and gram negative membranes. Using computational models of both membrane types previously made in the group, firstly, varying numbers of daptomycin molecules were placed randomly over the gram positive, specifically S. aureus, membrane and then run initially for 300ns. These simulations were continued, on Iridis 4, until the daptomycin molecules were no longer moving through the bacterial membrane at around 1500ns. These simulations have proposed that daptomycin moves slowly through the gram-positive membrane and only seem to disrupt the head groups of the phospholipids that make up the membrane. This could suggest that daptomycin does not need to enter the cell or completely destroy the membrane to cause cell death. This level of molecular detail is unobtainable experimentally, and whilst there are limitations to what these very short timescale simulations in relation to real time, it could give and insight into how daptomycin truly functions.
Currently, chlorhexidine simulations are running also on Iridis 4 having been set up similarly to the daptomycin simulations however both the gram positive and negative membranes are being examined. It is hoped that upon completion, these simulations will reveal the functionality of chlorhexidine and how it interacts with different membranes. This comparative study between two important and common antimicrobial agents, require a great deal of computational power due to the sheer number of simulations made but also the amount of time they are needed to run for to give meaningful results. Therefore, Iridis 4 has been vital to this work in order to achieve the time and size scales attempted to be replicated in this project.
Categories
Life sciences simulation: Biomolecular Organisation, Microbiology, Structural biology
Algorithms and computational methods: Molecular Dynamics
Simulation software: Gromacs
Visualisation and data handling software: VMD
Programming languages and libraries: Python