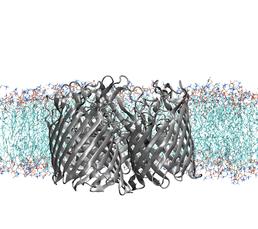
Membrane-Protein Interactions: The Outer Membrane of Gram-Negative Bacteria
- Started
- 10th June 2013
- Research Team
- Pin-Chia Hsu
- Investigators
- Syma Khalid
Understanding the interaction between membrane protein and biological membrane and predicting its interaction sites is one of the most important and key challenge research article in the modern structural biology. There is about 20-30% of membrane protein in genome and around 50% of membrane proteins were estimated and predicted as the future drug target. Furthermore, membrane proteins have both biological and biomedical importance, which involves multiple physiological functions in membrane, such as ion/small molecule transportation, energy transduction, cellular signalling and etc. Moreover, lipid-protein interactions are important in stabilizing its structure and in the native environment, and it helps the membrane proteins assembly.
Molecular dynamic simulation method provides a means of complement available structural, dynamic, thermodynamic information for protein embedded in a physiological, realistic, and modeled membrane environment. Using high performance computer, Iridis4, with it’s great facilities, will shorten the simulation time and could lead to study the variety of outer membrane proteins that share the similar structures and provide the better comparison results.
Categories
Life sciences simulation: Biomolecular simulations, Systems biology
Simulation software: Gromacs
Visualisation and data handling software: VMD, Xmgrace
Software Engineering Tools: Emacs
Programming languages and libraries: Fortran