Probing the oligomeric state and interaction surface of Fukutin Transmembrane Domain in lipid bilayer via Molecular Dynamics simulations
- Started
- 10th October 2010
- Research Team
- Daniel Holdbrook, Jamie Parkin, Nils Berglund, Yuk Leung
- Investigators
- Syma Khalid, Philip Williamson
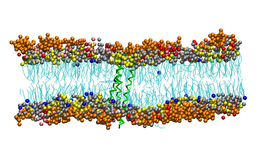
A side view of 3 FK1TMD (green) forming a trimer in a mix lipid bilayer. PIP2 headgroups are orange. Cholesterols are blue, PC are silver, PE are yellow, PS are pink and sphyngomyelin are red. All the lipid tails are in cyan.
Coarse-grained simulations revealed that FK1TMD readily formed dimers in DLPC bilayers with the tilt angle of the FK1TMD dimer is substantially reduced compared to an individual monomer, suggesting that the ability of the helices to tilt in the bilayer is affected by dimerisation.
The coarse-grain model was converted back to atomistic detail to further explore the conformational stability and dynamics of the FK1TMD dimer. The atomistic simulations showed that the most persistent interaction arose from the TXXSS motif in which hydrogen bonding was observed for up to 79% of the simulations between residues S17–S18 and S18–S18.
Finally, an attempt to discover how the lipid composition will affect the dimerisation properties of FK1TMD. CG simulations are set up with multiple FK1TMD proteins in a mixed lipid bilayer, which contains several different lipid moieties mimicking the in vivo Golgi membrane.
Categories
Life sciences simulation: Biomolecular simulations
Algorithms and computational methods: Molecular Dynamics
Simulation software: Gromacs
Visualisation and data handling software: VMD, Xmgrace
Computational platforms: Mac OS X