Biomolecular Organisation
For queries about this topic, contact Syma Khalid.
View the calendar of events relating to this topic.
Projects
Bioinformatic identification and physiological analysis of ethanol-related genes in C. elegans
Richard Edwards, Vincent O'Connor, Lindy Holden-Dye (Investigators), Ben Ient
Investigating the broad molecular, cellular and systems level impacts of acute and chronic ethanol in the nematode, Caenorhabditis elegans, as a model.
Cellular Automata Modelling of Membrane Formation and Protocell Evolution
Seth Bullock (Investigator), Stuart Bartlett
We simulated the meso-level behaviour of lipid-like particles in a range of chemical and physical environments. Self-organised protocellular structures can be shown to emerge spontaneously in systems with random, homogeneous initial conditions. Introducing an additional 'toxic' particle species and an associated set of synthesis reactions produced a new set of ecological behaviours compared to the original model of Ono and Ikegami.

Centre for Doctoral Training in Next Generation Computational Modelling
Hans Fangohr, Ian Hawke, Peter Horak (Investigators), Susanne Ufermann Fangohr, Thorsten Wittemeier, Ashley Setter, Kieran Selvon, Hossam Ragheb, Craig Rafter, Alvaro Perez-Diaz, Ryan Pepper, David Lusher, Stephen Gow, James Harrison, Paul Chambers, Jan Kamenik, Ioannis Begleris, Robert Entwistle, Jonathon Waters, Rory Brown, Joshua Greenhalgh, Emanuele Zappia
The £10million Centre for Doctoral Training was launched in November 2013 and is jointly funded by EPSRC, the University of Southampton, and its partners.
The NGCM brings together world-class simulation modelling research activities from across the University of Southampton and hosts a 4-year doctoral training programme that is the first of its kind in the UK.

Integrated in silico prediction of protein-protein interaction motifs
Richard Edwards (Investigator), Nicolas Palopoli, Kieren Lythgow
Many vital protein-protein interactions are mediated by Short Linear Motifs (SLiMs) which are short proteins typically 5-15 amino acids long containing only a few positions crucial to function. This project integrates a number of leading computational techniques to predict novel SLiMs and add crucial detail to protein-protein interaction networks.

Interactome-wide prediction of short linear protein interaction motifs in humans
Richard Edwards (Investigator)
Short Linear Motifs (SLiMs) are important in many protein-protein interactions. In previous work, we have developed a computational tool, SLiMFinder, which places the interpretation of evidence for motifs within a statistical framework with high specificity, and subsequently enhanced sensitivity through application of conservation-based sequence masking. We are now applying these tools to a comprehensive set of human protein-protein interactions in order to predict novel human SLiMs in silico.
Lyotropic phase transitions of lipids studied by CG MD simulation and experimental techniques
Syma Khalid (Investigator), Josephine Corsi
A study of the phase behaviour of cationic lipid - DNA complexes such as those used for transfection by coarse grained molecular dynamics simulation. Lipid systems studied include DOPE, DOPE/DNA and DOPE/DOTAP/DNA. Structural parameters and phase behaviour observed computationally have been compared with those gained using Small Angle X-ray Scattering (SAXS) and polarising light microscopy techniques.
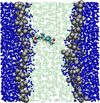
Multiscale modelling of biological membranes
Jonathan Essex (Investigator), Mario Orsi
Biological membranes are complex and fascinating systems, characterised by proteins floating in a sea of lipids. Biomembranes, besides being the fundamental structures employed by nature to encapsulate cells, play crucial roles in many phenomena indispensable for life, such as growth, energy storage, and in general information transduction via neural activity. In this project, we develop and apply multiscale computational models to simulate biological membranes and obtain molecular-level insights into fundamental structures and phenomena.
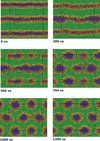
Simulation of biological systems at long length and distance scales
Jonathan Essex (Investigator), Kieran Selvon
This project aims to shed light on cell membrane mechanisms which are difficult to probe experimentally, in particular drug permiation across the cell membrane. If one had a full understanding of the mechanism, drugs could be designed to target particular embedded proteins to improve their efficacy, the viability of nano based medicines and materials could also be assessed, testing for toxicity etc.

Using Molecular Dynamics to Understand the Antibacterial Mechanisms of Daptomycin & Chlorhexidine to Target the Bacterial Membrane
This project aims to use molecular dynamics techniques to understand how antimicrobial peptides, daptomycin and chlorhexidine, disrupt both gram positive and negative cell membranes on an atomic level.

Using Molecular Dynamics to Understand the Antibacterial Mechanisms of Daptomycin & Chlorhexidine to Target the Bacterial Membrane
This project aims to use molecular dynamics techniques to understand how antimicrobial peptides, daptomycin and chlorhexidine, disrupt both gram positive and negative cell membranes on an atomic level.

Using Molecular Dynamics to Understand the Antibacterial Mechanisms of Daptomycin & Chlorhexidine to Target the Bacterial Membrane
Syma Khalid (Investigator), Eilish McBurnie
This project aims to use molecular dynamics techniques to understand how antimicrobial peptides, daptomycin and chlorhexidine, disrupt both gram positive and negative cell membranes on an atomic level.
People
 Seth Bullock
Seth BullockProfessor, Electronics and Computer Science (FPAS)
 Jonathan Essex
Jonathan EssexProfessor, Chemistry (FNES)
 Hans Fangohr
Hans FangohrProfessor, Engineering Sciences (FEE)
 Lindy Holden-Dye
Lindy Holden-DyeProfessor, Biological Sciences (FNES)
 Peter Horak
Peter HorakReader, Optoelectronics Research Centre
 Vincent O'Connor
Vincent O'ConnorReader, Biological Sciences (FNES)
 Tiina Roose
Tiina RooseReader, Engineering Sciences (FEE)
 Paul Skipp
Paul SkippReader, Biological Sciences (FNES)
 Robert Ewing
Robert EwingSenior Lecturer, Biological Sciences (FNES)
 Srinandan Dasmahapatra
Srinandan DasmahapatraLecturer, Electronics and Computer Science (FPAS)
 Ian Hawke
Ian HawkeLecturer, Mathematics (FSHS)
 Syma Khalid
Syma KhalidPrincipal Research Fellow, Chemistry (FNES)
 Philip Williamson
Philip WilliamsonSenior Research Fellow, Biological Sciences (FNES)
 Stuart Bartlett
Stuart BartlettPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Ioannis Begleris
Ioannis BeglerisPostgraduate Research Student, Engineering Sciences (FEE)
 Rory Brown
Rory BrownPostgraduate Research Student, Civil Engineering & the Environment (FEE)
 Paul Chambers
Paul ChambersPostgraduate Research Student, Engineering Sciences (FEE)
 Caroline Duignan
Caroline DuignanPostgraduate Research Student, Biological Sciences (FNES)
 Joseph Egan
Joseph EganPostgraduate Research Student, Mathematics (FSHS)
 Robert Entwistle
Robert EntwistlePostgraduate Research Student, Engineering Sciences (FEE)
 Ric Gillams
Ric GillamsPostgraduate Research Student, Chemistry (FNES)
 Stephen Gow
Stephen GowPostgraduate Research Student, Engineering Sciences (FEE)
 Joshua Greenhalgh
Joshua GreenhalghPostgraduate Research Student, Engineering Sciences (FEE)
 James Harrison
James HarrisonPostgraduate Research Student, Engineering Sciences (FEE)
 Tom Hebbron
Tom HebbronPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Harry L
Harry LPostgraduate Research Student, Biological Sciences (FNES)
 David Lusher
David LusherPostgraduate Research Student, Engineering Sciences (FEE)
 Alvaro Perez-Diaz
Alvaro Perez-DiazPostgraduate Research Student, Engineering Sciences (FEE)
 Can Pervane
Can PervanePostgraduate Research Student, Electronics and Computer Science (FPAS)
 Craig Rafter
Craig RafterPostgraduate Research Student, Engineering Sciences (FEE)
 Hossam Ragheb
Hossam RaghebPostgraduate Research Student, Engineering Sciences (FEE)
 Kieran Selvon
Kieran SelvonPostgraduate Research Student, Engineering Sciences (FEE)
 Ashley Setter
Ashley SetterPostgraduate Research Student, Engineering Sciences (FEE)
 Jonathon Waters
Jonathon WatersPostgraduate Research Student, Engineering Sciences (FEE)
 Thorsten Wittemeier
Thorsten WittemeierPostgraduate Research Student, Engineering Sciences (FEE)
 Emanuele Zappia
Emanuele ZappiaPostgraduate Research Student, Engineering Sciences (FEE)
 Matthew Higgins
Matthew HigginsUndergraduate Research Student, Biological Sciences (FNES)
 Elena Vataga
Elena VatagaTechnical Staff, iSolutions
 Petrina Butler
Petrina ButlerAdministrative Staff, Research and Innovation Services
 Susanne Ufermann Fangohr
Susanne Ufermann FangohrAdministrative Staff, Civil Engineering & the Environment (FEE)
 Josephine Corsi
Josephine CorsiAlumnus, University of Southampton
 Richard Edwards
Richard EdwardsAlumnus, University of New South Wales, Australia
 Ben Ient
Ben IentAlumnus, Biological Sciences (FNES)
 Jan Kamenik
Jan KamenikAlumnus, University of Southampton
 Kieren Lythgow
Kieren LythgowAlumnus, Health Protection Agency
 Nicolas Palopoli
Nicolas PalopoliAlumnus, Biological Sciences (FNES)
 Mario Orsi
Mario OrsiExternal Member, Queen Mary University of London
 Eilish McBurnie
Eilish McBurnieNone, None