Structural biology
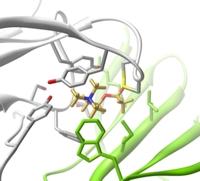
Structural biology is concerned with the physical and mechanical properties of biological molecules, including proteins, nucleotides, lipids, drugs and other small molecules. Topics include computational techniques for interpreting experimental data (e.g. NMR and X-Ray crystallography) as well as methods for predicting structure and function in the absence of experimental data (e.g. homology modelling and molecular dynamics).
Image courtesy: Phil Williamson
For queries about this topic, contact Philip Williamson.
View the calendar of events relating to this topic.
Projects

Centre for Doctoral Training in Next Generation Computational Modelling
Hans Fangohr, Ian Hawke, Peter Horak (Investigators), Susanne Ufermann Fangohr, Thorsten Wittemeier, Kieran Selvon, Alvaro Perez-Diaz, David Lusher, Ashley Setter, Emanuele Zappia, Hossam Ragheb, Ryan Pepper, Stephen Gow, Jan Kamenik, Paul Chambers, Robert Entwistle, Rory Brown, Joshua Greenhalgh, James Harrison, Jonathon Waters, Ioannis Begleris, Craig Rafter
The £10million Centre for Doctoral Training was launched in November 2013 and is jointly funded by EPSRC, the University of Southampton, and its partners.
The NGCM brings together world-class simulation modelling research activities from across the University of Southampton and hosts a 4-year doctoral training programme that is the first of its kind in the UK.

Control and Prediction of the Organic Solid State
Richard Boardman
This project aims to produce a computer technology for the prediction of the crystal structure(s) of an organic molecule, that could be used even prior to the synthesis of the compound.
Such a computational study could be done relatively quickly to predict the dangers and opportunities of the solid phases of a molecule under development. Our project will develop the methods of experimental screening for polymorphs and their characterisation, and hence the combination will provide a major new technology for aiding industrial formulation.
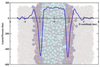
How far can we stretch the MARTINI?
Syma Khalid (Investigator), Ric Gillams
To date, coarse-grained lipid models have generally been parameterised to ensure the correct prediction of structural properties of membranes, such as the area per lipid and the bilayer thickness. The work described here explores the extent to which coarse-grained models are able to predict correctly bulk properties of lipids (phase behaviour) as well as the mechanical properties, such as lateral pressure profiles and stored elastic stress in bilayers. Such an evaluation is crucial for understanding the predictive capabilities of coarse-grained models.
Immunotherapy Research: Modelling MHC Class I Complex Assembly
Timothy Elliott, Jorn Werner (Investigators), Alistair Bailey
This project uses mathematical modelling and simulation to investigate mechanisms by which our cells process and present biological information that is used by our immune system to distinguish between healthy and diseased cells.

Integrated in silico prediction of protein-protein interaction motifs
Richard Edwards (Investigator), Nicolas Palopoli, Kieren Lythgow
Many vital protein-protein interactions are mediated by Short Linear Motifs (SLiMs) which are short proteins typically 5-15 amino acids long containing only a few positions crucial to function. This project integrates a number of leading computational techniques to predict novel SLiMs and add crucial detail to protein-protein interaction networks.
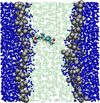
Multiscale modelling of biological membranes
Jonathan Essex (Investigator), Mario Orsi
Biological membranes are complex and fascinating systems, characterised by proteins floating in a sea of lipids. Biomembranes, besides being the fundamental structures employed by nature to encapsulate cells, play crucial roles in many phenomena indispensable for life, such as growth, energy storage, and in general information transduction via neural activity. In this project, we develop and apply multiscale computational models to simulate biological membranes and obtain molecular-level insights into fundamental structures and phenomena.

The application and critical assessment of protein-ligand binding affinities
Jonathan Essex (Investigator), Ioannis Haldoupis
A method that can accurately predict the binding affinity of small molecules to a protein target would be imperative to pharmaceutical development due to the time and resources that could be saved. A head-to-head comparison of such methodology, ranging from approximate methods to more rigorous methods, is performed in order to assess their accuracy and utility across a range of targets.
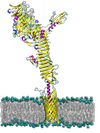
The autotransporter ? domain: insights into structure and function through multi scale molecular dynamics simulations
Syma Khalid (Investigator), Daniel Holdbrook, Thomas Piggot
We are performing a series of molecular dynamics simulations involving all autotransporters with known structure. We aim to identify key structural and dynamic properties in this family of proteins.

Using Molecular Dynamics to Understand the Antibacterial Mechanisms of Daptomycin & Chlorhexidine to Target the Bacterial Membrane
This project aims to use molecular dynamics techniques to understand how antimicrobial peptides, daptomycin and chlorhexidine, disrupt both gram positive and negative cell membranes on an atomic level.

Using Molecular Dynamics to Understand the Antibacterial Mechanisms of Daptomycin & Chlorhexidine to Target the Bacterial Membrane
This project aims to use molecular dynamics techniques to understand how antimicrobial peptides, daptomycin and chlorhexidine, disrupt both gram positive and negative cell membranes on an atomic level.

Using Molecular Dynamics to Understand the Antibacterial Mechanisms of Daptomycin & Chlorhexidine to Target the Bacterial Membrane
Syma Khalid (Investigator), Eilish McBurnie
This project aims to use molecular dynamics techniques to understand how antimicrobial peptides, daptomycin and chlorhexidine, disrupt both gram positive and negative cell membranes on an atomic level.
People
 Andrew Collins
Andrew CollinsProfessor, Medicine (FM)
 Timothy Elliott
Timothy ElliottProfessor, Medicine (FM)
 Jonathan Essex
Jonathan EssexProfessor, Chemistry (FNES)
 Hans Fangohr
Hans FangohrProfessor, Engineering Sciences (FEE)
 Anthony Postle
Anthony PostleProfessor, Medicine (FM)
 Peter Horak
Peter HorakReader, Optoelectronics Research Centre
 Tiina Roose
Tiina RooseReader, Engineering Sciences (FEE)
 Jorn Werner
Jorn WernerReader, Biological Sciences (FNES)
 Ian Hawke
Ian HawkeLecturer, Mathematics (FSHS)
 Syma Khalid
Syma KhalidPrincipal Research Fellow, Chemistry (FNES)
 Richard Boardman
Richard BoardmanSenior Research Fellow, Engineering Sciences (FEE)
 Philip Williamson
Philip WilliamsonSenior Research Fellow, Biological Sciences (FNES)
 Alistair Bailey
Alistair BaileyResearch Fellow, Medicine (FM)
 Gaia Andreoletti
Gaia AndreolettiPostgraduate Research Student, Medicine (FM)
 Ioannis Begleris
Ioannis BeglerisPostgraduate Research Student, Engineering Sciences (FEE)
 Louise Bolton
Louise BoltonPostgraduate Research Student, Medicine (FM)
 Rory Brown
Rory BrownPostgraduate Research Student, Civil Engineering & the Environment (FEE)
 Paul Chambers
Paul ChambersPostgraduate Research Student, Engineering Sciences (FEE)
 Paul Cross
Paul CrossPostgraduate Research Student, Engineering Sciences (FEE)
 Caroline Duignan
Caroline DuignanPostgraduate Research Student, Biological Sciences (FNES)
 Robert Entwistle
Robert EntwistlePostgraduate Research Student, Engineering Sciences (FEE)
 Ric Gillams
Ric GillamsPostgraduate Research Student, Chemistry (FNES)
 Stephen Gow
Stephen GowPostgraduate Research Student, Engineering Sciences (FEE)
 Joshua Greenhalgh
Joshua GreenhalghPostgraduate Research Student, Engineering Sciences (FEE)
 Ioannis Haldoupis
Ioannis HaldoupisPostgraduate Research Student, Chemistry (FNES)
 James Harrison
James HarrisonPostgraduate Research Student, Engineering Sciences (FEE)
 William Hurndall
William HurndallPostgraduate Research Student, Electronics and Computer Science (FPAS)
 David Lusher
David LusherPostgraduate Research Student, Engineering Sciences (FEE)
 Alvaro Perez-Diaz
Alvaro Perez-DiazPostgraduate Research Student, Engineering Sciences (FEE)
 Can Pervane
Can PervanePostgraduate Research Student, Electronics and Computer Science (FPAS)
 Craig Rafter
Craig RafterPostgraduate Research Student, Engineering Sciences (FEE)
 Hossam Ragheb
Hossam RaghebPostgraduate Research Student, Engineering Sciences (FEE)
 Kieran Selvon
Kieran SelvonPostgraduate Research Student, Engineering Sciences (FEE)
 Ashley Setter
Ashley SetterPostgraduate Research Student, Engineering Sciences (FEE)
 Jonathon Waters
Jonathon WatersPostgraduate Research Student, Engineering Sciences (FEE)
 Thorsten Wittemeier
Thorsten WittemeierPostgraduate Research Student, Engineering Sciences (FEE)
 Emanuele Zappia
Emanuele ZappiaPostgraduate Research Student, Engineering Sciences (FEE)
 Matthew Higgins
Matthew HigginsUndergraduate Research Student, Biological Sciences (FNES)
 Elena Vataga
Elena VatagaTechnical Staff, iSolutions
 Petrina Butler
Petrina ButlerAdministrative Staff, Research and Innovation Services
 Susanne Ufermann Fangohr
Susanne Ufermann FangohrAdministrative Staff, Civil Engineering & the Environment (FEE)
 Richard Edwards
Richard EdwardsAlumnus, University of New South Wales, Australia
 Jan Kamenik
Jan KamenikAlumnus, University of Southampton
 Kieren Lythgow
Kieren LythgowAlumnus, Health Protection Agency
 Nicolas Palopoli
Nicolas PalopoliAlumnus, Biological Sciences (FNES)
 Mario Orsi
Mario OrsiExternal Member, Queen Mary University of London
 Daniel Holdbrook
Daniel HoldbrookNone, None
 Eilish McBurnie
Eilish McBurnieNone, None
 Thomas Piggot
Thomas PiggotNone, None