Systems biology
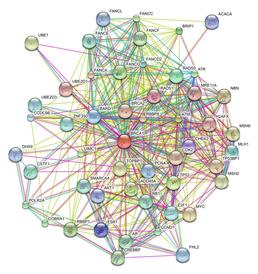
Systems Biology is the study of whole biological systems rather than individual components in isolation. The focus is on discovering emergent system properties that cannot be easily determined using a "sum of the parts" approach. Systems biology typically makes use of high throughput "Omics" technologies. This topic include projects modelling networks of interactions between genes, proteins, cells or whole organisms as well as the interaction between different levels of organisation.
For queries about this topic, contact Benjamin Macarthur.
View the calendar of events relating to this topic.
Projects

Amorphous Computation, Random Graphs and Complex Biological Networks
Seth Bullock (Investigator)
This interdisciplinary research collaboration arose within the Simple Models of Complex Networks research cluster funded by the EPSRC www.epsrca.ac.uk through the Novel Computation Initiative. Here, leading groups from the Universities of Leeds, Sheffield, Nottingham, Southampton, Royal Holloway and King’s College and industrial partners BT are brought together for the first time to develop novel amorphous computation methods based on the theory of random graphs.
Bioinformatic identification and physiological analysis of ethanol-related genes in C. elegans
Richard Edwards, Vincent O'Connor, Lindy Holden-Dye (Investigators), Ben Ient
Investigating the broad molecular, cellular and systems level impacts of acute and chronic ethanol in the nematode, Caenorhabditis elegans, as a model.

Centre for Doctoral Training in Next Generation Computational Modelling
Hans Fangohr, Ian Hawke, Peter Horak (Investigators), Susanne Ufermann Fangohr, Thorsten Wittemeier, Ashley Setter, Kieran Selvon, Hossam Ragheb, Craig Rafter, Alvaro Perez-Diaz, Ryan Pepper, David Lusher, Stephen Gow, James Harrison, Paul Chambers, Jan Kamenik, Ioannis Begleris, Robert Entwistle, Jonathon Waters, Rory Brown, Joshua Greenhalgh, Emanuele Zappia
The £10million Centre for Doctoral Training was launched in November 2013 and is jointly funded by EPSRC, the University of Southampton, and its partners.
The NGCM brings together world-class simulation modelling research activities from across the University of Southampton and hosts a 4-year doctoral training programme that is the first of its kind in the UK.
Development of wide-ranging functionality in ONETEP
Chris-Kriton Skylaris (Investigator), Jacek Dziedzic
ONETEP is at the cutting edge of developments in first principles calculations. However, while the fundamental difficulties of performing accurate first-principles calculations with linear-scaling cost have been solved, only a small core of functionality is currently available in ONETEP which prevents its wide application. In this collaborative project between three Universities, the original developers of ONETEP will lead an ambitious workplan whereby the functionality of the code will be rapidly and significantly enriched.
How far can we stretch the MARTINI?
Syma Khalid (Investigator), Ric Gillams
To date, coarse-grained lipid models have generally been parameterised to ensure the correct prediction of structural properties of membranes, such as the area per lipid and the bilayer thickness. The work described here explores the extent to which coarse-grained models are able to predict correctly bulk properties of lipids (phase behaviour) as well as the mechanical properties, such as lateral pressure profiles and stored elastic stress in bilayers. Such an evaluation is crucial for understanding the predictive capabilities of coarse-grained models.
Immunotherapy Research: Modelling MHC Class I Complex Assembly
Timothy Elliott, Jorn Werner (Investigators), Alistair Bailey
This project uses mathematical modelling and simulation to investigate mechanisms by which our cells process and present biological information that is used by our immune system to distinguish between healthy and diseased cells.

Integrated in silico prediction of protein-protein interaction motifs
Richard Edwards (Investigator), Nicolas Palopoli, Kieren Lythgow
Many vital protein-protein interactions are mediated by Short Linear Motifs (SLiMs) which are short proteins typically 5-15 amino acids long containing only a few positions crucial to function. This project integrates a number of leading computational techniques to predict novel SLiMs and add crucial detail to protein-protein interaction networks.

Interactome-wide prediction of short linear protein interaction motifs in humans
Richard Edwards (Investigator)
Short Linear Motifs (SLiMs) are important in many protein-protein interactions. In previous work, we have developed a computational tool, SLiMFinder, which places the interpretation of evidence for motifs within a statistical framework with high specificity, and subsequently enhanced sensitivity through application of conservation-based sequence masking. We are now applying these tools to a comprehensive set of human protein-protein interactions in order to predict novel human SLiMs in silico.
Mathematical modelling of plant nutrient uptake
Tiina Roose (Investigator)
In this project I will describe a model of plant water and nutrient uptake and how to translate this model and experimental data from the single root scale to the root branching structure scale.

Membrane-Protein Interactions: The Outer Membrane of Gram-Negative Bacteria
Syma Khalid (Investigator), Pin-Chia Hsu
The aim of the project is to looking for the interaction sites, which may responsible for turning on/off activity in outer membrane protein with gram-negative bacteria membrane using molecular dynamic (MD) approach.

Models of Avian Flocking
Edward Butler (Investigator)
This research review project explores current state of avian flock modelling research, exploring how ideas have developed since the earliest theories dating back to the 1930s.

Multiscale Modelling of Cellular Calcium Signalling
Hans Fangohr, Jonathan Essex (Investigators), Dan Mason
Calcium ions play a vitally important role in signal transduction and are key to many cellular processes including muscle contraction and cell apoptosis (cell death). This importance has made calcium an active area in biomedical science and mathematical modelling.
OMSys Towards a system model of a bacterial outer membrane
Syma Khalid (Investigator)
Many bacteria have an outer membrane which is the interface between the cell and its environment. The components of this membrane are well studied at an individual level, but there is a need to model and understand the outer membrane as a whole. In this project we aim to develop such a model of a bacterial outer membrane, linking computer simulations of the component molecules through to a more "systems biology" approach to modelling the outer membrane as a whole. Such an approach to modelling an OM must be multi-scale i.e. it must embrace a number of levels ranging from atomistic level modelling of e.g. the component proteins through to higher level "agent-based" modelling of the interplay of multiple components within the outer membrane as a whole. The different levels of description will be integrated to enable predictive modelling in order to explore the roles of outer membrane changes in e.g. antibiotic resistance.
Simulating Household Decision Making in Rural Malawi
James Dyke, Kate Schreckenberg (Investigators), Samantha Dobbie
A scoping exercise to determine whether data collection tools of the social sciences can be used effectively in the construction of empirical ABM. Focus fell upon simulating drought coping strategies of Malawian smallholders. Model implementation enabled inferences to be made concerning the impact of drought and input subsidies upon smallholder food security.

TEDx: Closing the Loop: Entropy Accounting for a Sustainable World
Stuart Bartlett (Investigator)
This is a TEDx talk that I gave on some ideas I've had about the large-scale thermodynamic organisation of life on Earth. While these ideas probably aren't new, I believe they can teach us something about the way in which we think about energy and the 'consumption' of goods and energy.
The ONETEP project
Chris-Kriton Skylaris (Investigator), Stephen Fox, Chris Pittock, Álvaro Ruiz-Serrano, Jacek Dziedzic
Program for large-scale quantum mechanical simulations of matter from first principles quantum mechanics. Based on theory and algorithms we have developed for linear-scaling density functional theory calculations on parallel computers.

The tarsal intersegmental reflex control system in the locust hind leg
David Simpson, Philip Newland (Investigators), Alicia Costalago Meruelo
Locomotion is vital for vertebrates and invertebrates to survive. Despite that feet are responsible for stability and agility in most animals, research on feet movements and their reflexes is scarce.
In this thesis, the tarsal reflex responses of locust will be studied and modelled with ANNs to achieve a deeper comprehension of how stability and agility is accomplished.
The choice of ANNs is linked to the applicability of the method into other fields, such as technological designs or medical treatment.
Tissue Engineering
Tiina Roose (Investigator)
This project deals with applying mathematical and computational modelling techniques to answer questions that are useful for tissue engineering applications.
Uncovering extensive post-translation regulation during human cell cycle progression by integrative multi-’omics analysis
Gregory Parkes (Investigator), Mahesan Niranjan
Analysis of high-throughput multi-’omics interactions across the hierarchy of expression has wide interest in making inferences with regard to biological function and biomarker discovery. Expression levels across different scales are determined by robust synthesis, regulation and degradation processes, and hence transcript (mRNA) measurements made by microarray/RNA-Seq only show modest correlation with corresponding protein levels.
In this work we are interested in quantitative modelling of correlation across such gene products. Building on recent work, we develop computational models spanning transcript, translation and protein levels at different stages of the H. sapiens cell cycle. We enhance this analysis by incorporating 25+ sequence-derived features which are likely determinants of cellular protein concentration and quantitatively select for relevant features, producing a vast dataset with thousands of genes. We reveal insights into the complex interplay between expression levels across time, using machine learning methods to highlight outliers with respect to such models as proteins associated with post-translationally regulated modes of action.
We uncover quantitative separation between modified and degraded proteins that have roles in cell cycle regulation, chromatin remodelling and protein catabolism according to Gene Ontology; and highlight the opportunities for providing biological insights in future model systems.

µ-VIS Computed Tomography Centre
Ian Sinclair, Richard Boardman, Dmitry Grinev, Philipp Thurner, Simon Cox, Jeremy Frey, Mark Spearing, Kenji Takeda (Investigators)
A dedicated centre for computed tomography (CT) at Southampton, providing complete support for 3D imaging science, serving Engineering, Biomedical, Environmental and Archaeological Sciences. The centre encompasses five complementary scanning systems supporting resolutions down to 200nm and imaging volumes in excess of one metre: from a matchstick to a tree trunk, from an ant's wing to a gas turbine blade.
People
 Jacek Brodzki
Jacek BrodzkiProfessor, Mathematics (FSHS)
 Seth Bullock
Seth BullockProfessor, Electronics and Computer Science (FPAS)
 Andrew Collins
Andrew CollinsProfessor, Medicine (FM)
 Simon Cox
Simon CoxProfessor, Engineering Sciences (FEE)
 Timothy Elliott
Timothy ElliottProfessor, Medicine (FM)
 Jonathan Essex
Jonathan EssexProfessor, Chemistry (FNES)
 Hans Fangohr
Hans FangohrProfessor, Engineering Sciences (FEE)
 Jeremy Frey
Jeremy FreyProfessor, Chemistry (FNES)
 Martin Glennie
Martin GlennieProfessor, Medicine (FM)
 Lindy Holden-Dye
Lindy Holden-DyeProfessor, Biological Sciences (FNES)
 Philip Newland
Philip NewlandProfessor, Biological Sciences (FNES)
 Graham Niblo
Graham NibloProfessor, Mathematics (FSHS)
 Mahesan Niranjan
Mahesan NiranjanProfessor, Electronics and Computer Science (FPAS)
 Anthony Postle
Anthony PostleProfessor, Medicine (FM)
 Ian Sinclair
Ian SinclairProfessor, Engineering Sciences (FEE)
 Peter Smith
Peter SmithProfessor, Institute for Life Sciences (FHS)
 Mark Spearing
Mark SpearingProfessor, Engineering Sciences (FEE)
 Karen Temple
Karen TempleProfessor, Medicine (FM)
 Peter Horak
Peter HorakReader, Optoelectronics Research Centre
 Rohan Lewis
Rohan LewisReader, Medicine (FM)
 Vincent O'Connor
Vincent O'ConnorReader, Biological Sciences (FNES)
 Tiina Roose
Tiina RooseReader, Engineering Sciences (FEE)
 Paul Skipp
Paul SkippReader, Biological Sciences (FNES)
 Ali Tavassoli
Ali TavassoliReader, Chemistry (FNES)
 Jorn Werner
Jorn WernerReader, Biological Sciences (FNES)
 Thomas Blumensath
Thomas BlumensathSenior Lecturer, Institute of Sound & Vibration Research (FEE)
 Robert Ewing
Robert EwingSenior Lecturer, Biological Sciences (FNES)
 David Simpson
David SimpsonSenior Lecturer, Institute of Sound & Vibration Research (FEE)
 Srinandan Dasmahapatra
Srinandan DasmahapatraLecturer, Electronics and Computer Science (FPAS)
 James Dyke
James DykeLecturer, Electronics and Computer Science (FPAS)
 Ian Hawke
Ian HawkeLecturer, Mathematics (FSHS)
 Kate Schreckenberg
Kate SchreckenbergLecturer, Civil Engineering & the Environment (FEE)
 Chris-Kriton Skylaris
Chris-Kriton SkylarisLecturer, Chemistry (FNES)
 Philipp Thurner
Philipp ThurnerLecturer, Engineering Sciences (FEE)
 Syma Khalid
Syma KhalidPrincipal Research Fellow, Chemistry (FNES)
 Richard Boardman
Richard BoardmanSenior Research Fellow, Engineering Sciences (FEE)
 Philip Williamson
Philip WilliamsonSenior Research Fellow, Biological Sciences (FNES)
 Alistair Bailey
Alistair BaileyResearch Fellow, Medicine (FM)
 Jacek Dziedzic
Jacek DziedzicResearch Fellow, Chemistry (FNES)
 Dmitry Grinev
Dmitry GrinevResearch Fellow, Engineering Sciences (FEE)
 Rob Mills
Rob MillsResearch Fellow, Electronics and Computer Science (FPAS)
 - -
- -Postgraduate Research Student, Electronics and Computer Science (FPAS)
 Jordi Arranz
Jordi ArranzPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Alice Ball
Alice BallPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Stuart Bartlett
Stuart BartlettPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Ioannis Begleris
Ioannis BeglerisPostgraduate Research Student, Engineering Sciences (FEE)
 Louise Bolton
Louise BoltonPostgraduate Research Student, Medicine (FM)
 Rory Brown
Rory BrownPostgraduate Research Student, Civil Engineering & the Environment (FEE)
 Edward Butler
Edward ButlerPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Paul Chambers
Paul ChambersPostgraduate Research Student, Engineering Sciences (FEE)
 Alicia Costalago Meruelo
Alicia Costalago MerueloPostgraduate Research Student, University of Southampton
 Samantha Dobbie
Samantha DobbiePostgraduate Research Student, Electronics and Computer Science (FPAS)
 Caroline Duignan
Caroline DuignanPostgraduate Research Student, Biological Sciences (FNES)
 Graham Elliott
Graham ElliottPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Robert Entwistle
Robert EntwistlePostgraduate Research Student, Engineering Sciences (FEE)
 Stephen Fox
Stephen FoxPostgraduate Research Student, Chemistry (FNES)
 Ric Gillams
Ric GillamsPostgraduate Research Student, Chemistry (FNES)
 Stephen Gow
Stephen GowPostgraduate Research Student, Engineering Sciences (FEE)
 Joshua Greenhalgh
Joshua GreenhalghPostgraduate Research Student, Engineering Sciences (FEE)
 James Harrison
James HarrisonPostgraduate Research Student, Engineering Sciences (FEE)
 Tom Hebbron
Tom HebbronPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Reza J. Forooshani
Reza J. ForooshaniPostgraduate Research Student, Medicine (FM)
 Guy Jacobs
Guy JacobsPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Harry L
Harry LPostgraduate Research Student, Biological Sciences (FNES)
 David Lusher
David LusherPostgraduate Research Student, Engineering Sciences (FEE)
 Gregory Parkes
Gregory ParkesPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Alvaro Perez-Diaz
Alvaro Perez-DiazPostgraduate Research Student, Engineering Sciences (FEE)
 Can Pervane
Can PervanePostgraduate Research Student, Electronics and Computer Science (FPAS)
 Maximillian Phipps
Maximillian PhippsPostgraduate Research Student, Chemistry (FNES)
 Chris Pittock
Chris PittockPostgraduate Research Student, Chemistry (FNES)
 Daniel Power
Daniel PowerPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Craig Rafter
Craig RafterPostgraduate Research Student, Engineering Sciences (FEE)
 Hossam Ragheb
Hossam RaghebPostgraduate Research Student, Engineering Sciences (FEE)
 Sonya Ridden
Sonya RiddenPostgraduate Research Student, Mathematics (FSHS)
 Álvaro Ruiz-Serrano
Álvaro Ruiz-SerranoPostgraduate Research Student, Chemistry (FNES)
 Kieran Selvon
Kieran SelvonPostgraduate Research Student, Engineering Sciences (FEE)
 Ashley Setter
Ashley SetterPostgraduate Research Student, Engineering Sciences (FEE)
 Jonathon Waters
Jonathon WatersPostgraduate Research Student, Engineering Sciences (FEE)
 Angela Watkins
Angela WatkinsPostgraduate Research Student, Biological Sciences (FNES)
 Thorsten Wittemeier
Thorsten WittemeierPostgraduate Research Student, Engineering Sciences (FEE)
 Emanuele Zappia
Emanuele ZappiaPostgraduate Research Student, Engineering Sciences (FEE)
 Elena Vataga
Elena VatagaTechnical Staff, iSolutions
 Petrina Butler
Petrina ButlerAdministrative Staff, Research and Innovation Services
 Susanne Ufermann Fangohr
Susanne Ufermann FangohrAdministrative Staff, Civil Engineering & the Environment (FEE)
 Ella Marley-Zagar
Ella Marley-ZagarEnterprise staff, Medicine (FM)
 Richard Edwards
Richard EdwardsAlumnus, University of New South Wales, Australia
 Ben Ient
Ben IentAlumnus, Biological Sciences (FNES)
 Jan Kamenik
Jan KamenikAlumnus, University of Southampton
 Kieren Lythgow
Kieren LythgowAlumnus, Health Protection Agency
 Dan Mason
Dan MasonAlumnus, University of Southampton
 Nicolas Palopoli
Nicolas PalopoliAlumnus, Biological Sciences (FNES)
 Joe Scutt Phillips
Joe Scutt PhillipsAlumnus, Centre for the Environment, Fisheries and Aquaculture Sciences
 Kenji Takeda
Kenji TakedaAlumnus, Engineering Sciences (FEE)
 Brian Bonney
Brian BonneyNone, None
 Pin-Chia Hsu
Pin-Chia HsuNone, None