Biomedical
The Biomedical topic covers all projects modelling medical or disease processes in order to develop or improve treatments and therapeutics. This topic includes rational drug design, clinical trials and modelling the molecular basis of disease, inlcuding genetic and epigenetic contributions.
For queries about this topic, contact Richard Edwards.
View the calendar of events relating to this topic.
Projects

A Mathematical Analysis of the Driving Force of Perivascular Drainage in the Brain
Giles Richardson, Roxana-Octavia Carare (Investigators), Alexandra Diem
The observation that solute drainage in the brain occurs in the reverse direction of the blood flow has for a long time been puzzling for researchers. We developed a simple analytical model that can explain this reverse drainage of solutes and has potential implications for the development of treatment for Alzheimer's Disease.
Application of RNA-Seq for gene fusion identification in blood cancers
William Tapper (Investigator), Marcin Knut
Gene fusions are often the cause of different blood cancers. As such, accurate identification of them provides information on the underlying cause of a cancer, ensuring appropriate choice of treatment. However, due to shortcomings of the currently applied methods for gene fusion identification, some of them escape undetected. We are employing RNA-Seq, a cutting-edge method for sequencing RNA, the messenger of genetic information, to investigate gene fusions.
Bioinformatic identification and physiological analysis of ethanol-related genes in C. elegans
Richard Edwards, Vincent O'Connor, Lindy Holden-Dye (Investigators), Ben Ient
Investigating the broad molecular, cellular and systems level impacts of acute and chronic ethanol in the nematode, Caenorhabditis elegans, as a model.

Centre for Doctoral Training in Next Generation Computational Modelling
Hans Fangohr, Ian Hawke, Peter Horak (Investigators), Susanne Ufermann Fangohr, Thorsten Wittemeier, Kieran Selvon, Alvaro Perez-Diaz, David Lusher, Ashley Setter, Emanuele Zappia, Hossam Ragheb, Ryan Pepper, Stephen Gow, Jan Kamenik, Paul Chambers, Robert Entwistle, Rory Brown, Joshua Greenhalgh, James Harrison, Jonathon Waters, Ioannis Begleris, Craig Rafter
The £10million Centre for Doctoral Training was launched in November 2013 and is jointly funded by EPSRC, the University of Southampton, and its partners.
The NGCM brings together world-class simulation modelling research activities from across the University of Southampton and hosts a 4-year doctoral training programme that is the first of its kind in the UK.

Coronary Artery Stent Design for Challenging Disease
Neil Bressloff (Investigator), Georgios Ragkousis
In this work, a method has been setup to (i) reconstruct diseased patient specific coronary artery segments; (ii) use the new supercomputer to run many simulations of this complex problem and (iii) assess the degree of stent malapposition. The aim now is to devise a stent delivery system that can mitigate this problem
DePuy Technology Partnership
Mark Taylor (Investigator), Adam Briscoe
This initiative concerns the transfer of knowledge between three key institutions (University of Southampton, University of Leeds and University of Hamburg) and DePuy International limited. The project is concerned with the ongoing advancement of technology used in orthopaedic devices.
Identification of Gene-Modules Associated to a Predisposition of Post-Traumatic Stress Disorder
Christopher Woelk (Investigator), Michael Breen
The predisposing genetic factors associated to Post-Traumatic Stress Disorder (PTSD) are altogether unknown. Since not all trauma-survivors later develop the PTSD, it has been hypothesized that transcript differentiation prior–to-trauma exposure could be associated to the risk and resilience of PTSD. We apply a systems-level approach to investigate changes in transcript abundance (gene expression profiles) in whole blood of U.S. Marines sampled prior-to-deployment to the battlefield and followed through-out a seven month deployment to obtain disorder related outcomes.
Identification of novel Crustacean Pathogen Receptor Proteins
Richard Edwards, Chris Hauton, Timothy Elliott (Investigators), Oyindamola Lawal, Lloyd Mushambadzi
We are mining EST libraries (sequence fragments of expressed genes) for novel proteins that might play a role in the immune response of crustaceans.
Identification of phage DNA, common insertion sites and their effect on genes within S.pneumoniae
Richard Edwards, Amy Dean
This study seeks to find if there are any common insertion sites across different strains of S.pneumoniae and discover genes that undergo frequent mutation due to phages and if these mutations can be linked to virulence of the strains.
Identifying factors required for DNA methylation using the imprinting control protein ZFP57
Deborah Mackay (Investigator)
Mutation of ZFP57 in humans is associated with widespread loss of DNA methylation at imprinted genes, and clinical features including congenital anomalies and developmental delay (Mackay et al, 08). This indicates that ZFP57 is required for DNA methylation of imprinted genes necessary for normal development.
We propose to identify the DNA sequences targeted by ZFP57, and its protein cofactors. This work will give insight into the biology of imprinting, indicate mechanisms of disease, and identify novel imprinted genes.
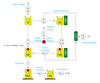
Immunotherapy Research: Modelling MHC Class I Complex Assembly
Timothy Elliott, Jorn Werner (Investigators), Alistair Bailey
This project uses mathematical modelling and simulation to investigate mechanisms by which our cells process and present biological information that is used by our immune system to distinguish between healthy and diseased cells.
.small.png)
Imprinting Disorders Finding Out Why
Karen Temple (Investigator)
We are conducting a research project to determine the cause and clinical impact of widespread imprinting aberrations in human development. We are recruiting patients with possible or definite imprinting disorders (due to methylation loss or gain at an imprinted loci)
including Silver Russell syndrome, Transient Neonatal diabetes, Beckwith Wiedemann syndrome, Angelman syndrome Prader Willi syndrome, UPD 14 syndromes and Pseudohypoparathyroidism.

Integrated in silico prediction of protein-protein interaction motifs
Richard Edwards (Investigator), Nicolas Palopoli, Kieren Lythgow
Many vital protein-protein interactions are mediated by Short Linear Motifs (SLiMs) which are short proteins typically 5-15 amino acids long containing only a few positions crucial to function. This project integrates a number of leading computational techniques to predict novel SLiMs and add crucial detail to protein-protein interaction networks.

Interactome-wide prediction of short linear protein interaction motifs in humans
Richard Edwards (Investigator)
Short Linear Motifs (SLiMs) are important in many protein-protein interactions. In previous work, we have developed a computational tool, SLiMFinder, which places the interpretation of evidence for motifs within a statistical framework with high specificity, and subsequently enhanced sensitivity through application of conservation-based sequence masking. We are now applying these tools to a comprehensive set of human protein-protein interactions in order to predict novel human SLiMs in silico.

Measuring biomolecules - improvements to the spectroscopic ruler
Pavlos Lagoudakis, Tom Brown (Investigators), Jan Junis Rindermann, James Richardson
The spectroscopic ruler is a technique to measure the geometry of biomolecules on the nm scale by labeling them with pairs of fluorescent markers and measuring distance dependent non-radiative energy transfer between them. The remaining uncertainty in the application of the technique originates from the unknown orientation between the optical dipole moments of the fluorescent markers, especially when the molecule undergoes thermal fluctuations in physiological conditions. Recently we introduced a simulation based method for the interpretation of the fluorescence decay dynamics of the markers that allows us to retrieve both the average orientation and the extent of directional fluctuations of the involved dipole moments.

Microstructural modeling of skin mechanics
Georges Limbert (Investigator), Emanuele Zappia
Microstructural modeling of skin mechanics to gain a mechanistic insight into the biomechanics of the skin.

Modelling neuronal activity at the knee joint
Mark Taylor, Tiina Roose (Investigators), Gwen Palmer
The function of the knee joint is reliant on proprioception, which involves the response of nerve endings in the tissues at the joint. This project will be concentrating on the neuronal activity, caused by mechanical stimuli, of the more common receptors found at the knee (Ruffini, Paciniform, Golgi and Nociceptor).
There are three stages to this project:
1. Modelling the behaviour of each individual receptor, with the use of the Hodgkin-Huxley model [1].
2. These models will then be applied to the soft tissues around a knee, where a global deformation of the tissue will result in local stimulation of receptors.
3. The soft tissue models will then be applied to structures in the knee.
[1] - Hodgkin, A.L. and A.F. Huxley, A quantitative description of membrane current and its application to conduction and excitation in nerve. Journal of Physiology, 1952. 117: p. 500-544.
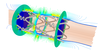
Multi-objective design optimisation of coronary stents
Neil Bressloff, Georges Limbert (Investigators), Sanjay Pant
Stents are tubular type scaffolds that are deployed (using an inflatable balloon on a catheter), most commonly to recover the shape of narrowed (diseased) arterial segments. Despite the widespread clinical use of stents in cardiovascular intervention, the presence of such devices can cause adverse responses leading to fatality or to the need for further treatment. The most common unwanted responses of inflammation are in-stent restenosis and thrombosis. Such adverse biological responses in a stented artery are influenced by many factors, including the design of the stent. This project aims at using multi-objective optimisation techniques to find an optimum family of coronary stents which are more resistant to the processes of in-stent restenosis (IR) and stent thrombosis (ST).
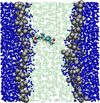
Multiscale modelling of biological membranes
Jonathan Essex (Investigator), Mario Orsi
Biological membranes are complex and fascinating systems, characterised by proteins floating in a sea of lipids. Biomembranes, besides being the fundamental structures employed by nature to encapsulate cells, play crucial roles in many phenomena indispensable for life, such as growth, energy storage, and in general information transduction via neural activity. In this project, we develop and apply multiscale computational models to simulate biological membranes and obtain molecular-level insights into fundamental structures and phenomena.

Multiscale models of magnetic materials at elevated temperatures
Denis Kramer (Investigator), Jonathon Waters
This project will develop and apply multi-scale modelling approaches to investigate thermal fluctuation effects in magnetic materials.

MXL Project
Mark Taylor, Junfen Shi (Investigators)
‘MXL’ is short for “Enhanced patient safety by computational Modelling from clinically available X-rays to minimise the risk of overload and instability for optimised function and Longevity”. This is an international EU-funded project which the Bioengineering Sciences Research Group at Southampton is involved in. For more information, visit http://www.m-x-l.eu

Perceived Attractiveness as a Factor Affecting Condomless Sex
Anastasia Eleftheriou, Seth Bullock
The objectives of this project are to better understand the relationship between perceived attractiveness and condom use intentions and to gain insight into the relationship between perceived attractiveness and potential sexual risk behaviours.

Preventing Alzheimer's Disease: A Multiphysics Simulation Approach
Neil Bressloff, Giles Richardson, Roxana-Octavia Carare (Investigators), Alexandra Diem
Experimental research has identified the causes of many diseases, such as Alzheimer's Disease. However, finding an effective treatment is very cost- and time-intensive and sacrifices many animals and does not guarantee success. In this PhD project, we investigate the driving force of solute drainage in the brain using multiphysics simulations in order to identify possible ways of preventing dementia.
Respiratory mask modeling
Jacques Ernes
Abaqus modelling of repiratory masks, bioengineering, Health sciences

Sample tracking in whole-exome sequencing projects
Andrew Collins, Sarah Ennis (Investigators), Reuben Pengelly
Whole-exome sequencing is entering clinical use for genetic investigations, and it is therefore essential that robust quality control is utilised. As such we designed and validated a tool to allow for unambiguous tying of patient data to a patient, to identify, and thus prevent errors such as the switching of samples during processing.
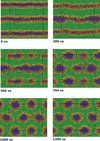
Simulation of biological systems at long length and distance scales
Jonathan Essex (Investigator), Kieran Selvon
This project aims to shed light on cell membrane mechanisms which are difficult to probe experimentally, in particular drug permiation across the cell membrane. If one had a full understanding of the mechanism, drugs could be designed to target particular embedded proteins to improve their efficacy, the viability of nano based medicines and materials could also be assessed, testing for toxicity etc.
Statistical model of the knee
Mark Taylor (Investigator), Francis Galloway, Prasanth Nair
Development of methods for large scale computational testing of a tibial tray incorporating inter-patient variability.

The application of next-generation sequencing to unresolved familial disease
Andrew Collins, Sarah Ennis (Investigators), Jane Gibson, Reuben Pengelly
Next-generation sequencing (NGS) allows us to sequence individual patients cost-effectively, allowing us to enter a new era of genomic medicine. The level of genetic detail that we can access through these methods is unprecedented making it suitable for clinical molecular diagnostics.

The tarsal intersegmental reflex control system in the locust hind leg
David Simpson, Philip Newland (Investigators), Alicia Costalago Meruelo
Locomotion is vital for vertebrates and invertebrates to survive. Despite that feet are responsible for stability and agility in most animals, research on feet movements and their reflexes is scarce.
In this thesis, the tarsal reflex responses of locust will be studied and modelled with ANNs to achieve a deeper comprehension of how stability and agility is accomplished.
The choice of ANNs is linked to the applicability of the method into other fields, such as technological designs or medical treatment.
Tissue Engineering
Tiina Roose (Investigator)
This project deals with applying mathematical and computational modelling techniques to answer questions that are useful for tissue engineering applications.

Transgenerational inheritance of allergy in a multi generational cohort
John Holloway (Investigator)
The aim of this project is to determine the vertical transmission of DNA methylation by identification of CpG sites by microarray analysis of 450,000 CpG sites in 252 women of the IoW cohort that are associated with allergic sensitization and testing whether the identified methylation patterns are vertically transmitted to their offspring and whether modifiable environmental conditions during gestation affect DNA methylation.
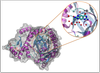
Water molecules in drug development: can we predict drug affinity when water molecules are involved?
Jonathan Essex (Investigator), Hannah Bruce Macdonald, Christopher Cave-Ayland
Water molecules are often found to be involved in drug-protein binding and can influence the effectiveness of a drug. We aim to aid drug design by calculating the energies involved with complexes of drugs, proteins and water molecules to predict the affinities of drug molecules.

µ-VIS Computed Tomography Centre
Ian Sinclair, Richard Boardman, Dmitry Grinev, Philipp Thurner, Simon Cox, Jeremy Frey, Mark Spearing, Kenji Takeda (Investigators)
A dedicated centre for computed tomography (CT) at Southampton, providing complete support for 3D imaging science, serving Engineering, Biomedical, Environmental and Archaeological Sciences. The centre encompasses five complementary scanning systems supporting resolutions down to 200nm and imaging volumes in excess of one metre: from a matchstick to a tree trunk, from an ant's wing to a gas turbine blade.
People
 Neil Bressloff
Neil BressloffProfessor, Engineering Sciences (FEE)
 Tom Brown
Tom BrownProfessor, Chemistry (FNES)
 Seth Bullock
Seth BullockProfessor, Electronics and Computer Science (FPAS)
 Andrew Collins
Andrew CollinsProfessor, Medicine (FM)
 Simon Cox
Simon CoxProfessor, Engineering Sciences (FEE)
 Timothy Elliott
Timothy ElliottProfessor, Medicine (FM)
 Sarah Ennis
Sarah EnnisProfessor, Medicine (FM)
 Jonathan Essex
Jonathan EssexProfessor, Chemistry (FNES)
 Hans Fangohr
Hans FangohrProfessor, Engineering Sciences (FEE)
 Jeremy Frey
Jeremy FreyProfessor, Chemistry (FNES)
 Lindy Holden-Dye
Lindy Holden-DyeProfessor, Biological Sciences (FNES)
 John Holloway
John HollowayProfessor, Medicine (FM)
 Pavlos Lagoudakis
Pavlos LagoudakisProfessor, Physics & Astronomy (FPAS)
 Philip Newland
Philip NewlandProfessor, Biological Sciences (FNES)
 Anthony Postle
Anthony PostleProfessor, Medicine (FM)
 Ian Sinclair
Ian SinclairProfessor, Engineering Sciences (FEE)
 Mark Spearing
Mark SpearingProfessor, Engineering Sciences (FEE)
 Karen Temple
Karen TempleProfessor, Medicine (FM)
 Peter Horak
Peter HorakReader, Optoelectronics Research Centre
 Rohan Lewis
Rohan LewisReader, Medicine (FM)
 Deborah Mackay
Deborah MackayReader, Medicine (FM)
 Vincent O'Connor
Vincent O'ConnorReader, Biological Sciences (FNES)
 Giles Richardson
Giles RichardsonReader, Mathematics (FSHS)
 Tiina Roose
Tiina RooseReader, Engineering Sciences (FEE)
 Paul Skipp
Paul SkippReader, Biological Sciences (FNES)
 Jorn Werner
Jorn WernerReader, Biological Sciences (FNES)
 Thomas Blumensath
Thomas BlumensathSenior Lecturer, Institute of Sound & Vibration Research (FEE)
 Roxana-Octavia Carare
Roxana-Octavia CarareSenior Lecturer, Medicine (FM)
 Robert Ewing
Robert EwingSenior Lecturer, Biological Sciences (FNES)
 Prasanth Nair
Prasanth NairSenior Lecturer, Engineering Sciences (FEE)
 Reuben Pengelly
Reuben PengellySenior Lecturer, Medicine (FM)
 David Simpson
David SimpsonSenior Lecturer, Institute of Sound & Vibration Research (FEE)
 Christopher Bell
Christopher BellLecturer, Biological Sciences (FNES)
 Jane Gibson
Jane GibsonLecturer, Biological Sciences (FNES)
 Ian Hawke
Ian HawkeLecturer, Mathematics (FSHS)
 Denis Kramer
Denis KramerLecturer, Engineering Sciences (FEE)
 Georges Limbert
Georges LimbertLecturer, Engineering Sciences (FEE)
 Philipp Thurner
Philipp ThurnerLecturer, Engineering Sciences (FEE)
 Richard Boardman
Richard BoardmanSenior Research Fellow, Engineering Sciences (FEE)
 Chris Hauton
Chris HautonSenior Research Fellow, Ocean & Earth Science (FNES)
 Philip Williamson
Philip WilliamsonSenior Research Fellow, Biological Sciences (FNES)
 Alistair Bailey
Alistair BaileyResearch Fellow, Medicine (FM)
 Andrea Boghi
Andrea BoghiResearch Fellow, Engineering Sciences (FEE)
 Adam Briscoe
Adam BriscoeResearch Fellow, Engineering Sciences (FEE)
 Dario Carugo
Dario CarugoResearch Fellow, Engineering Sciences (FEE)
 Dmitry Grinev
Dmitry GrinevResearch Fellow, Engineering Sciences (FEE)
 Gwen Palmer
Gwen PalmerResearch Fellow, Engineering Sciences (FEE)
 James Richardson
James RichardsonResearch Fellow, Chemistry (FNES)
 Roxana Aldea
Roxana AldeaPostgraduate Research Student, Mathematics (FSHS)
 Ioannis Begleris
Ioannis BeglerisPostgraduate Research Student, Engineering Sciences (FEE)
 Louise Bolton
Louise BoltonPostgraduate Research Student, Medicine (FM)
 Michael Breen
Michael BreenPostgraduate Research Student, Medicine (FM)
 Rory Brown
Rory BrownPostgraduate Research Student, Civil Engineering & the Environment (FEE)
 Hannah Bruce Macdonald
Hannah Bruce MacdonaldPostgraduate Research Student, Chemistry (FNES)
 Christopher Cave-Ayland
Christopher Cave-AylandPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Paul Chambers
Paul ChambersPostgraduate Research Student, Engineering Sciences (FEE)
 Dmitri Chernyshenko
Dmitri ChernyshenkoPostgraduate Research Student, Engineering Sciences (FEE)
 Michael Chesnaye
Michael ChesnayePostgraduate Research Student, Institute of Sound & Vibration Research (FEE)
 Alicia Costalago Meruelo
Alicia Costalago MerueloPostgraduate Research Student, University of Southampton
 Alexandra Diem
Alexandra DiemPostgraduate Research Student, Engineering Sciences (FEE)
 Joseph Egan
Joseph EganPostgraduate Research Student, Mathematics (FSHS)
 Anastasia Eleftheriou
Anastasia EleftheriouPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Robert Entwistle
Robert EntwistlePostgraduate Research Student, Engineering Sciences (FEE)
 Francis Galloway
Francis GallowayPostgraduate Research Student, Engineering Sciences (FEE)
 Ric Gillams
Ric GillamsPostgraduate Research Student, Chemistry (FNES)
 Stephen Gow
Stephen GowPostgraduate Research Student, Engineering Sciences (FEE)
 Joshua Greenhalgh
Joshua GreenhalghPostgraduate Research Student, Engineering Sciences (FEE)
 James Harrison
James HarrisonPostgraduate Research Student, Engineering Sciences (FEE)
 Tom Hebbron
Tom HebbronPostgraduate Research Student, Electronics and Computer Science (FPAS)
 Marcin Knut
Marcin KnutPostgraduate Research Student, Medicine (FM)
 David Lusher
David LusherPostgraduate Research Student, Engineering Sciences (FEE)
 Sanjay Pant
Sanjay PantPostgraduate Research Student, Engineering Sciences (FEE)
 Alvaro Perez-Diaz
Alvaro Perez-DiazPostgraduate Research Student, Engineering Sciences (FEE)
 Craig Rafter
Craig RafterPostgraduate Research Student, Engineering Sciences (FEE)
 Hossam Ragheb
Hossam RaghebPostgraduate Research Student, Engineering Sciences (FEE)
 Georgios Ragkousis
Georgios RagkousisPostgraduate Research Student, Engineering Sciences (FEE)
 Sonya Ridden
Sonya RiddenPostgraduate Research Student, Mathematics (FSHS)
 Jan Junis Rindermann
Jan Junis RindermannPostgraduate Research Student, Physics & Astronomy (FPAS)
 Kieran Selvon
Kieran SelvonPostgraduate Research Student, Engineering Sciences (FEE)
 Ashley Setter
Ashley SetterPostgraduate Research Student, Engineering Sciences (FEE)
 Jonathon Waters
Jonathon WatersPostgraduate Research Student, Engineering Sciences (FEE)
 Thorsten Wittemeier
Thorsten WittemeierPostgraduate Research Student, Engineering Sciences (FEE)
 Emanuele Zappia
Emanuele ZappiaPostgraduate Research Student, Engineering Sciences (FEE)
 Matthew Higgins
Matthew HigginsUndergraduate Research Student, Biological Sciences (FNES)
 Elena Vataga
Elena VatagaTechnical Staff, iSolutions
 Petrina Butler
Petrina ButlerAdministrative Staff, Research and Innovation Services
 Susanne Ufermann Fangohr
Susanne Ufermann FangohrAdministrative Staff, Civil Engineering & the Environment (FEE)
 Amy Dean
Amy DeanAlumnus, former UG, Biological Sciences
 Richard Edwards
Richard EdwardsAlumnus, University of New South Wales, Australia
 Ben Ient
Ben IentAlumnus, Biological Sciences (FNES)
 Jan Kamenik
Jan KamenikAlumnus, University of Southampton
 Kieren Lythgow
Kieren LythgowAlumnus, Health Protection Agency
 Lloyd Mushambadzi
Lloyd MushambadziAlumnus, former UG, Biological Sciences
 Nicolas Palopoli
Nicolas PalopoliAlumnus, Biological Sciences (FNES)
 Barbara Sander
Barbara SanderAlumnus, Chemistry (FNES)
 Kenji Takeda
Kenji TakedaAlumnus, Engineering Sciences (FEE)
 Jacques Ernes
Jacques ErnesExternal Member, Technical University of Eindhoven
 Mario Orsi
Mario OrsiExternal Member, Queen Mary University of London
 Zehong Au
Zehong AuNone, None
 Stephen Green
Stephen GreenNone, None
 Junfen Shi
Junfen ShiNone, None
 William Tapper
William TapperNone, None
 Christopher Woelk
Christopher WoelkNone, None